Introduction
The Pre2Med X-Protal (hereinafter referred to as Pre²MedX) is a comprehensive academic bioinformatics web server designed to support cancer precision therapy research. The platform enables transcriptomics-based prediction of drug response, identification of drug-sensitive and drug-resistant cellular subpopulations, prioritization of candidate anticancer drugs and potential combination strategies, and inference of immunotherapy response potential from both single-cell and bulk RNA sequencing data.
Platform Mission
To bridge the gap between genomic data and clinical applications by providing accessible, accurate, and interpretable pharmacological predictions for personalized medicine approaches in oncology and beyond.
Platform Overview
Pre2MedX integrates multiple published open-source algorithms into a unified analytical workflow, maintaining original code implementations under their respective open-source licenses while providing a standardized interface for academic research applications.
Platform Architecture
- Frontend Interface: Responsive web application with intuitive workflow design
- Analysis Engine: Dockerized algorithm containers with standardized I/O
- Data Management: Secure encrypted storage with automated data lifecycle management
- Compute Infrastructure: Scalable cloud-based processing with job queue management
- Visualization Module: Interactive result exploration with publication-quality graphics
Key Platform Features
Comprehensive Analysis Capabilities
- Drug Response Prediction: Quantify sensitivity/resistance patterns for individual drugs
- Subpopulation Identification: Discover pharmacologically distinct cell clusters within tumors
- Therapeutic Optimization: Recommend optimal single-agent or combination therapies
- Toxicity Assessment: Evaluate potential side effects on healthy cell populations
- Immunotherapy Suitability: Assess tumor immunogenicity and T-cell infiltration patterns
- Mechanistic Insights: Link pharmacological responses to molecular pathways and signatures
Integrated Algorithms
Pre2MedX integrates multiple validated algorithms for comprehensive pharmacogenomic analysis. All integrated methods are published, peer-reviewed academic tools; Pre²Med X-Portal integrates them within a standardized workflow.
scPharm
Computational framework designed to identify drug-sensitive and resistant cell subpopulations within tumors by integrating pharmacological profiles with single-cell RNA-sequencing data.
CaDRReS-Sc
Single-cell adaptation of the CaDRReS framework, developed for predicting drug response from transcriptomic data at the single-cell level.
scDEAL
Deep learning-based approach for predicting drug response outcomes using single-cell gene expression data.
Beyondcell
Computational framework for assessing therapeutic vulnerabilities and deriving drug sensitivity scores from single-cell transcriptomic data.
DREEP
Method for estimating drug response potentials directly from single-cell expression profiles.
CaDRReS
Method for predicting drug response and repositioning opportunities from bulk RNA-seq data.
oncoPredict
Comprehensive computational framework for predicting drug sensitivity in cancer genomics based on gene expression features.
pRRophetic
Algorithm for predicting clinical chemotherapeutic response from tumor gene expression levels.
CM-Drug
Signature based model for predicting effective combination therapies by analyzing immune checkpoint modulation
TIDE
Tumor Immune Dysfunction and Exclusion framework for immunotherapy response prediction.
Tres
Model for assessing T cell fitness to predict immunotherapy response across diverse tumor types.
Algorithm Licensing Note
All integrated algorithms are implemented under their original open-source licenses. Pre2MedX does not modify or redistribute original source code beyond necessary integration adaptations. Users are responsible for compliance with individual algorithm licensing terms for their specific applications.
Analysis Workflow
Step 1: Data Submission
- Authentication: Email-based login system for secure task management
- Job Configuration: Select algorithms, parameters, and analysis scope
- Data Upload: Secure encrypted transfer of sequencing data
- Validation: Automated quality control and format verification
Step 2: Processing Pipeline
- Queue Management: Automated job scheduling with priority handling
- Parallel Execution: Simultaneous algorithm execution when applicable
- Quality Monitoring: Real-time progress tracking and error detection
- Resource Optimization: Dynamic allocation based on analysis complexity
Step 3: Result Generation
- Data Integration: Consolidation of outputs from multiple algorithms
- Visualization: Generation of interactive and static result views
- Report Compilation: Comprehensive analysis summaries
- Notification: Email alerts upon completion or failure
Input Data Formats
Supported Data Formats
- scRNA-seq:
- Seurat Objects (.rds): Preferred format for single-cell analysis
- 10X Genomics H5: Standard 10X Genomics output format
- Expression Matrices: Gene × cell counts matrices, tab-delimited (TXT, as shown in the figure below)

- Bulk RNA-seq:
- Matrices Objects (.rds): Data frame format for expression analysis
- Expression Matrices: Gene × cell TPM/FPKM matrices, tab-delimited (TXT)
Data Requirements
Pre2MedX is optimized for tumor cell analysis. Input data should originate from tumor cell lines or clinical tumor samples. For single-cell data, we recommend preprocessing including quality control, normalization, and clustering prior to submission for optimal pharmacological analysis.
Parameter Configuration
Basic Parameters
- Upload Data File: Required. Upload a data file in txt, rds, or h5 format.
- File Format: Required. Select the format of the uploaded file:
- txt: Universal format for expression matrices (tab-delimited).
- rds: For Seurat objects or bulk RNA‑seq expression matrices.
- h5: Supports single‑cell expression profiles (e.g., 10X Genomics).
- Data Type: Required. Choose scRNA‑seq or bulk RNA‑seq; determines available methods.
- Task Name: Required. Assign a custom name for task identification.
- Run Immunotherapy: Required. Whether to perform immunotherapy response prediction.
- Cell Source: Required. Select Tumor Tissue or tumor Cell Line.
- Drug of Interest: Optional. Specify a drug name for focused analysis; if empty, all drugs are considered.
Method‑Specific Parameters
- CaDRReS Parameters:
- GDSC Version: Required. GDSC data version, default GDSC1.
- oncoPredict Parameters:
- Reference Pharmacology Dataset: Required. Choose CTRP2, GDSC1, or GDSC2.
- pRRophetic Parameters:
- Tissue Type: Required. Tissue type (e.g., all, breast, lung), default all.
- scPharm Parameters:
- Mode: Required. Operating mode: Single, Comb, or Both.
- DR analysis: Optional. Perform drug‑recommendation ranking.
- Combo analysis: Optional. Predict drug combinations.
- DSE analysis: Optional. Estimate drug side effects on normal cells (requires Tumor Tissue).
- scDEAL Parameters:
- Type: Optional. Use saved training nodes, default 1 (enabled).
- Beyondcell Parameters:
- Pathway Include: Optional. Include pathway signatures, default TRUE.
- CaDRReS‑Sc Parameters:
- GDSC Version: Required. GDSC data version, default GDSC1.
- DREEP Parameters:
- Pharmacological Data Selection: Required. Choose CTRP2, GDSC, or PRISM.
Additional Parameters (Optional)
The following optional parameters are available for each method after clicking the “More Params” button.
- scPharm Additional Parameters:
- Number of Components: PCA component count. Default: 30.
- Number of Genes (CELL-ID): Drug-related feature genes used. Default: 200.
- Number of CPU Cores: CPU cores allocated. Default: 1.
- Top N for Combo Analysis: Top drugs for combination analysis. Default: 5.
- Feature Names: Specify genes (comma-separated). Default: all genes.
- Slot of Seurat Object: Data slot (counts, data, scale.data). Default: data.
- Assay of Seurat Object: Assay type (e.g., RNA, SCT). Default: RNA.
- Sensitive Cell Threshold: Threshold for sensitive cells. Default: -1.751302.
- Resistant Cell Threshold: Threshold for resistant cells. Default: 1.518551.
- Beyondcell Additional Parameters:
- Clustering Resolution: Drug reaction cluster resolution. Default: 0.2.
- PCs: PCA dimensions for UMAP. Default: 10.
- CaDRReS-Sc Additional Parameters:
- Cancer Type: TCGA cancer type abbreviation. Default: (all).
- Cluster Mode: Label for clustering. Default: each cell as a separate cluster.
- DREEP Additional Parameters:
- Cluster Resolution: Clustering resolution. Default: 0.01.
- CaDRReS Additional Parameters:
- Cancer Type: TCGA cancer type abbreviation. Default: (all).
Parameter Optimization
Detailed parameter descriptions and optimization guidelines are available in our comprehensive documentation. For most applications, default parameters provide robust performance. Advanced users can fine-tune algorithm-specific parameters for specialized applications.
Accessing Results
Result Notification System
- Email Alerts: Immediate notification upon job completion or failure
- Task ID: Unique identifier for each analysis job
- Status Dashboard: Real-time monitoring of all submitted tasks
- Result Retrieval: Multiple access methods for convenience
Access Methods
- Task ID Search: Direct access using the unique task ID
- Email-based Retrieval: View all tasks associated with an email address
- User Dashboard: Registered users can access their complete analysis history
Visualization Interface
The result visualization page provides interactive exploration of analysis outputs:
- Pharmacological Landscapes: Visual representation of drug response patterns
- Subpopulation Analysis: Identification and characterization of sensitive/resistant clusters
- Drug Recommendation Plot: Visualization of ranked drug response predictions
- Combination Networks: Synergy scores for drug pairs
- Export Capabilities: Download images, tables, and complete analysis packages
Data Retention Policy
Due to storage resource limitations, analysis results are retained for 7 days after completion. Users should download and archive important results promptly. For privacy and security, we implement automatic data purging after the retention period.
Interpreting Results
The analysis results comprise an overview page featuring static chart panels covering sample descriptions, clustering and dimensionality reduction, predictions from various algorithms, and immunotherapy response. Users can click on panel titles to access interactive result pages, enabling more detailed exploration of the findings.
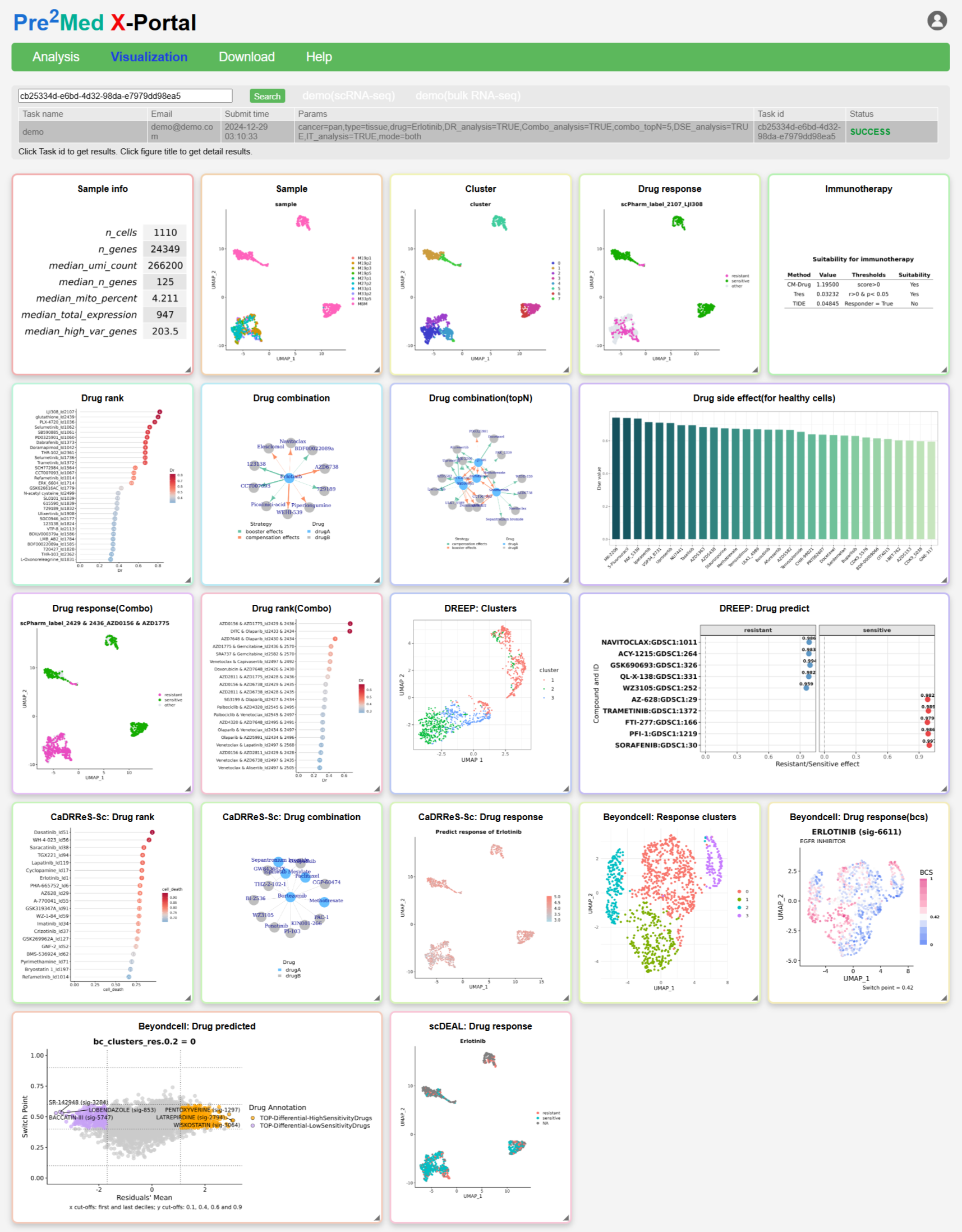
Result panel contains:
-
Sample Info
Summary statistics of the input single-cell dataset, including cell number, gene number, UMI counts, gene counts, mitochondrial percentage, and overall expression levels. -
Sample (UMAP)
UMAP visualization of cells. When multiple samples are provided, batch information is shown. Click to enter the interactive page for metadata, gene expression, and drug prediction queries. -
Cluster
Cell clustering results based on expression profiles, displayed on a UMAP embedding. Click to explore clusters, batches, gene expression, and drug responses interactively. -
scPharm: Drug Response
Single-cell–level prediction of drug sensitivity and resistance for a selected or top-ranked drug. Click to explore predicted responses across cells and clusters. -
Immunotherapy
Immune checkpoint therapy potential inferred using the TIDE, Tres and CM-Drug algorithms. -
scPharm: Drug Rank
Prioritized drug list generated by Drug Rank (DR) analysis. Higher-ranked drugs are predicted to be more effective for the sample. -
scPharm: Drug Combination
Predicted combination therapies for the top-ranked drug, including Booster and Compensation effects. Click to view the interactive drug combination network. -
scPharm: Drug Combination (Top N)
Drug combination predictions for the top N ranked drugs. Click to explore interactive combination networks of varying complexity. -
scPharm: Drug Side Effect (Healthy Cells)
Drug toxicity prediction for normal cells identified by CopyKAT. Higher-ranked drugs indicate stronger potential adverse effects. -
scPharm: Drug Response (Combo)
Single-cell drug response prediction in Combo or Both mode, based on GDSC combination drug data. -
scPharm: Drug Rank (Combo)
Recommended drug combinations ranked using GDSC combination data. -
DREEP: Clusters
UMAP clustering results from DREEP analysis. Differences from baseline UMAP are expected due to GF-ICF normalisation. -
DREEP: Drug Predict
Top sensitive and resistant drugs predicted by DREEP. Click to view all predicted drugs in an interactive scatter plot. -
CaDRReS-Sc: Drug Rank
Drug ranking based on predicted tumour cell death proportion at the single-cell level. -
CaDRReS-Sc: Drug Combination
Network visualization of top-ranked drug combinations predicted by CaDRReS-Sc. -
CaDRReS-Sc: Drug Response
Single-cell drug response prediction (LN_IC50) for a selected or top-ranked drug. -
Beyondcell: Response Clusters
Dimensionality reduction of cells based on Beyondcell Scores, highlighting clusters with similar drug response patterns. -
Beyondcell: Drug Response (BCS)
Calibrated Beyondcell Scores indicating cellular drug sensitivity. -
Beyondcell: Drug Predicted
Scatter plot of predicted drug responses categorised as sensitive, resistant, or others. -
scDEAL: Drug Response
Drug response prediction by scDEAL. Undefined cells reflect stringent quality control during scDEAL analysis.

Result panel contains:
-
Sample Info
Summary statistics of the input bulk dataset, including sample number, gene number, and median total expression levels. -
Immunotherapy
Immune checkpoint therapy potential inferred using the TIDE, Tres and CM-Drug algorithms. -
Sample (PCA)
PCA dimensionality reduction of bulk RNA-seq samples, showing global expression similarity among samples. -
Cluster
Hierarchical clustering dendrogram based on bulk expression profiles. -
CaDRReS: Drug Response
Drug response prediction (LN_IC50) for bulk RNA-seq data. Click to view predicted responses for all drugs in a heatmap. -
oncoPredict: Drug Response
Bulk RNA-seq drug response prediction (LN_IC50) using oncoPredict. Results are displayed as an interactive heatmap. -
pRRophetic: Drug Response
Drug response prediction (LN_IC50) for bulk RNA-seq data using pRRophetic.
For detailed results descriptions and examples, please check the Full Help Document.
Security and Privacy
Data Protection Measures
- Encrypted Storage: All user data encrypted at rest using AES-256
- Secure Transfer: TLS 1.3 encryption for all data transmissions
- Access Control: Role-based permissions and authentication requirements
- Data Isolation: Logical separation between user datasets
- Audit Logging: Comprehensive tracking of all data access events
Privacy Commitments
- No Data Sharing: User data is never shared with third parties
- Limited Retention: Automatic deletion after analysis completion
- Transparent Processing: Clear documentation of all analytical steps
- Academic Focus: Platform designed exclusively for research applications
Account Security
We recommend using email-based authentication for enhanced security. This approach allows password changes without losing access to analysis history and provides reliable notification delivery for job status updates.
Citation Policy
When using Pre2MedX for research publications, please cite both the platform and the specific algorithms employed in your analysis.
Platform Citation
Pre2Med X-Portal: Zheng, J., Chen, S., Cheng, Q., & Wang, H. (2025). Pre2Med X-Portal: An integrated web server for predictive and precision drug-response analysis. Pre2Med X-Portal. https://Pre2medx.org
Algorithm-Specific Citations
For scPharm drug response prediction:
Tian, P., Zheng, J., Qiao, K., Fan, Y., Xu, Y., Wu, T., Chen, S., Zhang, Y., Zhang, B., Ambrogio, C., & Wang, H. (2025). scPharm: Identifying Pharmacological Subpopulations of Single Cells for Precision Medicine in Cancers. Advanced Science, 12, e2412419. https://doi.org/10.1002/advs.202412419
For scDEAL drug response prediction:
Chen, J., Wang, X., Ma, A., Wang, Q.-E., Liu, B., Li, L., Xu, D., & Ma, Q. (2022). Deep transfer learning of cancer drug responses by integrating bulk and single-cell RNA-seq data. Nature Communications, 13, 6494. https://doi.org/10.1038/s41467-022-34277-7
For Beyondcell drug response prediction:
Fustero-Torre, C., Jiménez-Santos, M.J., García-Martín, S., Carretero-Puche, C., García-Jimeno, L., Ivanchuk, V., Di Domenico, T., Gómez-López, G., & Al-Shahrour, F. (2021). Beyondcell: targeting cancer therapeutic heterogeneity in single-cell RNA-seq data. Genome Medicine, 13, 187. https://doi.org/10.1186/s13073-021-01001-x
For CaDRReS-Sc drug response prediction:
Suphavilai, C., Chia, S., Sharma, A., Tu, L., Da Silva, R.P., Mongia, A., DasGupta, R., & Nagarajan, N. (2021). Predicting heterogeneity in clone-specific therapeutic vulnerabilities using single-cell transcriptomic signatures. Genome Medicine, 13, 189. https://doi.org/10.1186/s13073-021-01000-y
For DREEP drug response prediction:
Pellecchia, S., Viscido, G., Franchini, M., & Gambardella, G. (2023). Predicting drug response from single-cell expression profiles of tumours. BMC Medicine, 21, 476. https://doi.org/10.1186/s12916-023-03182-1
For oncoPredict drug response prediction:
Maeser, D., Gruener, R.F., & Huang, R.S. (2021). oncoPredict: an R package for predicting in vivo or cancer patient drug response and biomarkers from cell line screening data. Briefings in Bioinformatics, 22, bbab260. https://doi.org/10.1093/bib/bbab260
For pRRophetic drug response prediction:
Geeleher, P., Cox, N., & Huang, R.S. (2014). pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS ONE, 9, e107468. https://doi.org/10.1371/journal.pone.0107468
For CaDRReS drug response prediction:
Suphavilai, C., Bertrand, D., & Nagarajan, N. (2018). Predicting Cancer Drug Response using a Recommender System. Bioinformatics, 34, 3907-3914. https://doi.org/10.1093/bioinformatics/bty452
Immunotherapy Citations
For TIDE immunotherapy prediction:
Jiang, P., Gu, S., Pan, D., Fu, J., Sahu, A., Hu, X., Li, Z., Traugh, N., Bu, X., Li, B., Liu, J., Freeman, G. J., Brown, M. A., Wucherpfennig, K. W., & Liu, X. S. (2018). Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nature Medicine, 24, 1550–1558. https://doi.org/10.1038/s41591-018-0136-1
For Tres immunotherapy prediction:
Zhang, Y., Vu, T., Palmer, D. C., Wolf, N. K., Wang, Y. Z., Thaxton, J. E., Rhodes, C., Bevins, D., Drake, J., Innamarato, P., Park, S., Li, D., Hall, M., Gao, J., Wang, E., Weber, J. S., & Restifo, N. P. (2022). A T cell resilience model associated with response to immunotherapy in multiple tumor types. Nature Medicine, 28, 1421–1431. https://doi.org/10.1038/s41591-022-01799-y
For CM-Drug immunotherapy drug recommendation:
Xia, Y., Li, X., Bie, N., Pan, W., Miao, Y.-R., Yang, M., Gao, Y., Chen, C., Liu, H., Gan, L., & Guo, A.-Y. (2024). A method for predicting drugs that can boost the efficacy of immune checkpoint blockade. Nature Immunology, 25, 659-670. https://doi.org/10.1038/s41590-024-01789-x
Additional Citations
Please consult individual algorithm documentation for specific citation requirements. The full help documentation includes complete bibliographic references for all integrated methods.
Contact Information
We welcome feedback, suggestions, bug reports, and collaboration inquiries from the research community.
Institutional Address
School of Life Sciences and Technology
Tongji University
No. 1239, Siping Road, Yangpu District
Shanghai 200092, China
Telephone
Office: +86-21-65980233
Availability: Monday-Friday, 9:00-17:00 (GMT+8)